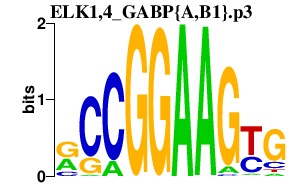
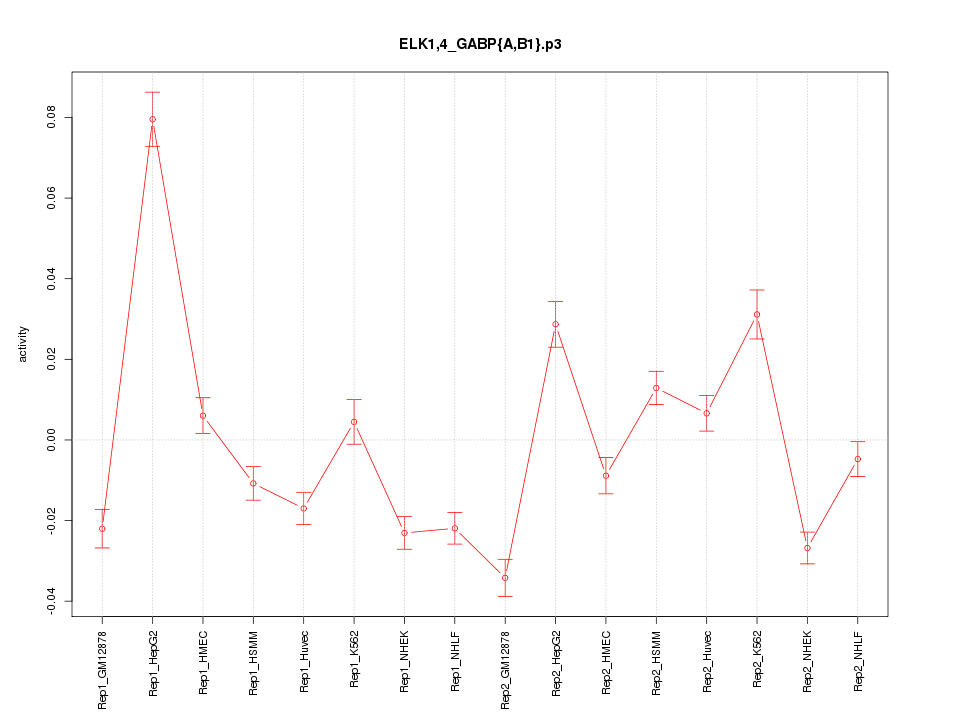
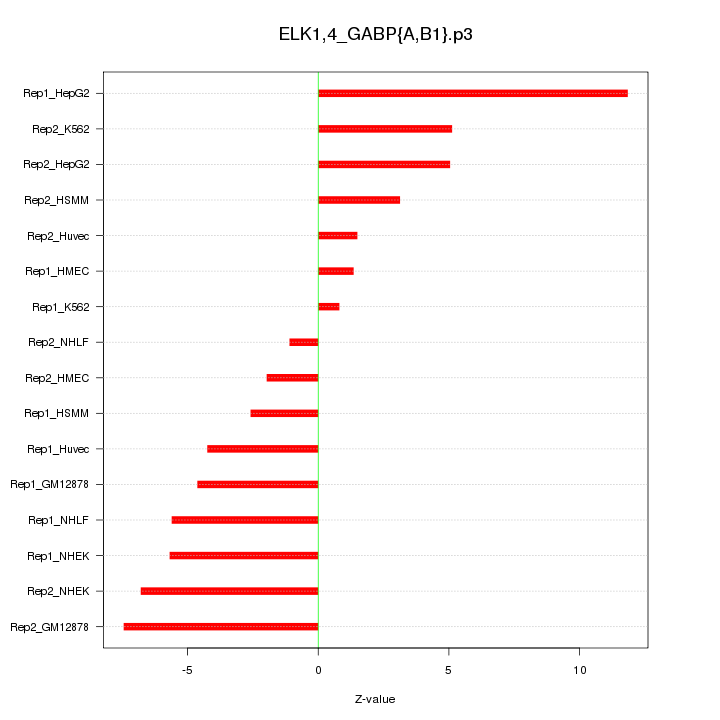
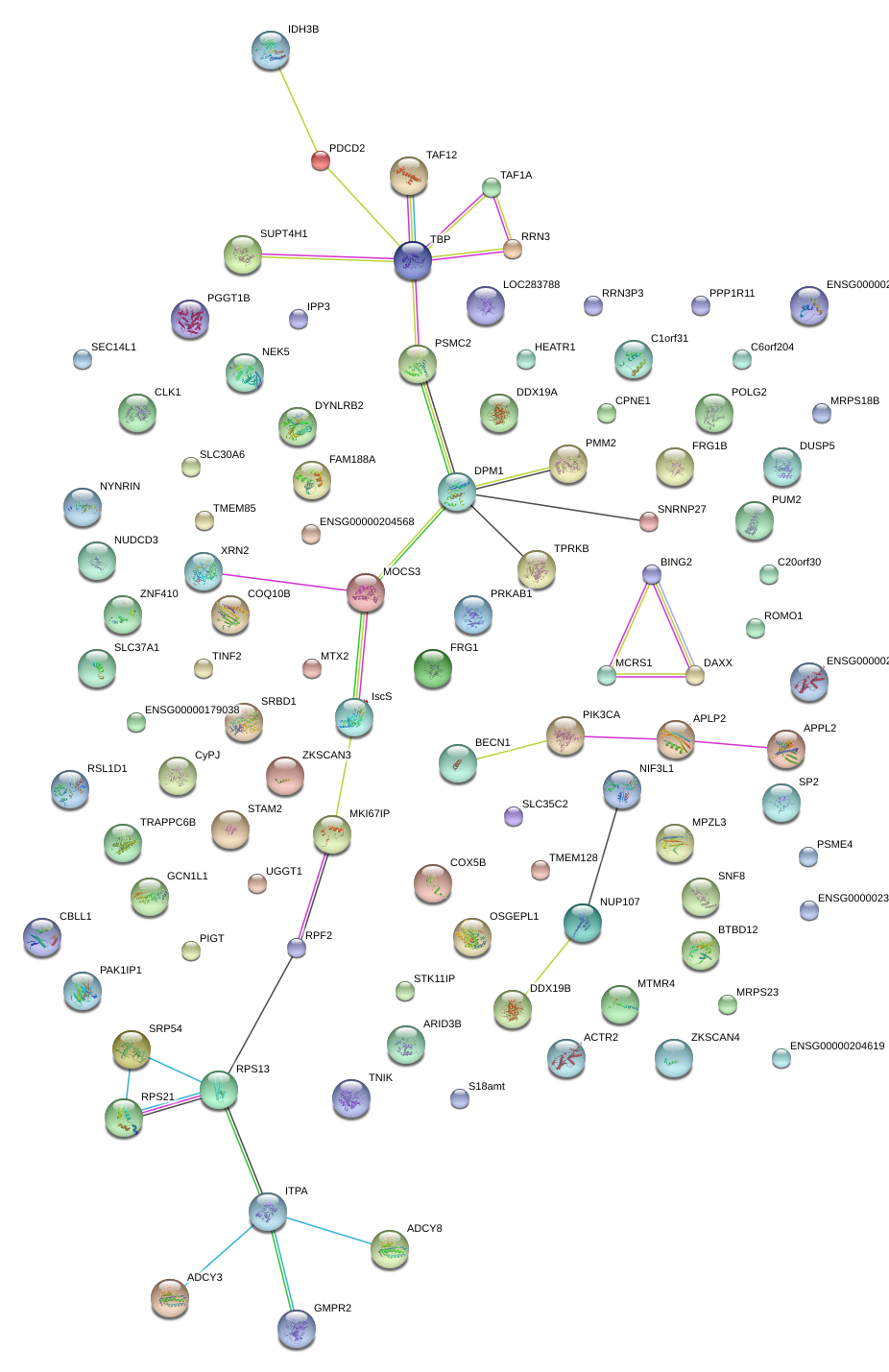
|
chr20_-_49008445
|
11.083
|
NM_003859
|
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit
|
|
chr2_-_152740387
|
9.254
|
NM_005843
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr20_+_21231921
|
7.617
|
NM_012255
|
XRN2
|
5'-3' exoribonuclease 2
|
|
chr20_+_49008754
|
7.061
|
NM_014484
|
MOCS3
|
molybdenum cofactor synthesis 3
|
|
chr20_+_60395515
|
6.791
|
NM_001024
|
RPS21
|
ribosomal protein S21
|
|
chr6_+_111409933
|
6.577
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr2_-_27147995
|
6.265
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr2_-_73817935
|
6.239
|
NM_016058
|
TPRKB
|
TP53RK binding protein
|
|
chr12_+_118590039
|
6.063
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr2_+_220170804
|
5.727
|
NM_052902
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr1_-_28842103
|
5.498
|
NM_001135218
NM_005644
|
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa
|
|
chr17_-_53784494
|
5.439
|
NM_003168
|
SUPT4H1
|
suppressor of Ty 4 homolog 1 (S. cerevisiae)
|
|
chr6_+_30693464
|
5.355
|
NM_014046
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr2_+_176842368
|
5.343
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr2_-_20413838
|
5.272
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr2_-_201461945
|
5.268
|
NM_032472
NM_130906
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3
|
|
chr2_+_32244406
|
5.168
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr3_-_130600931
|
5.138
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr2_-_54051290
|
5.113
|
NM_014614
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4
|
|
chr1_+_232575850
|
4.996
|
NM_001012985
|
C1orf31
|
chromosome 1 open reading frame 31
|
|
chr14_-_23781607
|
4.920
|
NM_001099274
NM_012461
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr21_-_29286982
|
4.916
|
NM_015565
|
LTN1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr6_-_119137923
|
4.915
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr12_-_104153920
|
4.886
|
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr11_-_17055763
|
4.847
|
NM_001017
|
RPS13
|
ribosomal protein S13
|
|
chr17_-_53282376
|
4.839
|
NM_016070
|
MRPS23
|
mitochondrial ribosomal protein S23
|
|
chr16_-_11852862
|
4.805
|
NM_015659
|
RSL1D1
|
ribosomal L1 domain containing 1
|
|
chr12_-_48248132
|
4.780
|
NM_006337
|
MCRS1
|
microspherule protein 1
|
|
chr14_-_23781328
|
4.755
|
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr20_+_60395623
|
4.694
|
|
RPS21
|
ribosomal protein S21
|
|
chr4_-_4300821
|
4.679
|
NM_032927
|
TMEM128
|
transmembrane protein 128
|
|
chr15_+_72620570
|
4.655
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr1_+_22224571
|
4.638
|
|
HSPC157
|
hypothetical LOC29092
|
|
chr17_-_53950191
|
4.622
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr17_-_44377094
|
4.619
|
NM_007241
|
SNF8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae)
|
|
chr2_+_201462294
|
4.615
|
NM_001136039
NM_001142355
NM_021824
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr12_-_119116848
|
4.607
|
NM_006836
|
GCN1L1
|
GCN1 general control of amino-acid synthesis 1-like 1 (yeast)
|
|
chr7_+_107171809
|
4.581
|
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr20_-_5041482
|
4.573
|
NM_001009923
NM_001009924
NM_001009925
NM_014145
|
C20orf30
|
chromosome 20 open reading frame 30
|
|
chr20_-_44426407
|
4.555
|
NM_015945
NM_173073
NM_173179
|
SLC35C2
|
solute carrier family 35, member C2
|
|
chr20_+_18496068
|
4.525
|
|
LOC388789
|
hypothetical LOC388789
|
|
chr2_+_97628929
|
4.511
|
NM_001862
|
COX5B
|
cytochrome c oxidase subunit Vb
|
|
chr15_+_32304477
|
4.478
|
NM_016454
|
TMEM85
|
transmembrane protein 85
|
|
chr16_+_68938264
|
4.467
|
NM_018332
|
DDX19A
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19A
|
|
chr14_+_34521945
|
4.424
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr14_-_38709161
|
4.358
|
NM_001079537
NM_177452
|
TRAPPC6B
|
trafficking protein particle complex 6B
|
|
chr16_+_21926939
|
4.354
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr20_+_43478120
|
4.353
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr10_-_15942461
|
4.271
|
NM_024948
|
FAM188A
|
family with sequence similarity 188, member A
|
|
chr20_-_2592777
|
4.270
|
NM_006899
NM_174855
NM_174856
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr7_-_44496703
|
4.225
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr6_+_170705345
|
4.210
|
NM_001172085
NM_003194
|
TBP
|
TATA box binding protein
|
|
chr20_+_3138055
|
4.208
|
NM_033453
NM_181493
|
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase)
|
|
chr14_+_34521854
|
4.172
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr21_+_42807052
|
4.157
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr20_+_33750645
|
4.126
|
NM_080748
|
ROMO1
|
reactive oxygen species modulator 1
|
|
chr6_-_28327980
|
4.119
|
NM_019110
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4
|
|
chr16_+_79132354
|
4.100
|
NM_130897
|
DYNLRB2
|
dynein, light chain, roadblock-type 2
|
|
chr16_-_3601580
|
4.057
|
NM_032444
|
SLX4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae)
|
|
chr14_+_23937766
|
4.044
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr6_+_10803141
|
4.017
|
NM_017906
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr14_+_73423260
|
3.978
|
NM_021188
|
ZNF410
|
zinc finger protein 410
|
|
chr2_-_122210873
|
3.934
|
NM_032390
|
MKI67IP
|
MKI67 (FHA domain) interacting nucleolar phosphoprotein
|
|
chr10_+_112247614
|
3.906
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr2_+_128565269
|
3.883
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr16_-_15095590
|
3.875
|
NM_018427
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae)
|
|
chr16_+_8799170
|
3.833
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr20_+_28225511
|
3.829
|
|
LOC283788
FRG1B
|
FSHD region gene 1 pseudogene
FSHD region gene 1 family, member B
|
|
chr17_-_59923547
|
3.816
|
NM_007215
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit
|
|
chr14_+_23771464
|
3.816
|
NM_001002000
NM_001002001
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr5_-_114626422
|
3.750
|
NM_005023
|
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr1_-_220829704
|
3.725
|
NM_005681
NM_139352
|
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa
|
|
chr16_+_2593351
|
3.685
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr6_+_30142910
|
3.652
|
NM_021959
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11
|
|
chr17_+_43328514
|
3.635
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr6_-_33398656
|
3.622
|
NM_001141969
NM_001141970
NM_001350
|
DAXX
|
death-domain associated protein
|
|
chr2_-_24995402
|
3.622
|
NM_004036
|
ADCY3
|
adenylate cyclase 3
|
|
chr17_+_72596349
|
3.612
|
|
SEC14L1
C17orf86
|
SEC14-like 1 (S. cerevisiae)
chromosome 17 open reading frame 86
|
|
chr3_+_180348818
|
3.611
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr2_-_74915722
|
3.610
|
|
|
|
|
chr2_+_198026470
|
3.607
|
NM_025147
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae)
|
|
chr12_+_67366989
|
3.594
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr17_-_38229790
|
3.558
|
NM_003766
|
BECN1
|
beclin 1, autophagy related
|
|
chr20_-_33750457
|
3.549
|
NM_001198989
NM_021100
|
NFS1
|
NFS1 nitrogen fixation 1 homolog (S. cerevisiae)
|
|
chr2_+_69974581
|
3.544
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr6_-_170735593
|
3.494
|
NM_002598
NM_144781
|
PDCD2
|
programmed cell death 2
|
|
chr2_+_65308332
|
3.487
|
NM_001005386
NM_005722
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast)
|
|
chr7_+_102775309
|
3.483
|
NM_002803
|
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2
|
|
chr1_-_234834410
|
3.482
|
NM_018072
|
HEATR1
|
HEAT repeat containing 1
|
|
chr2_-_190336168
|
3.464
|
NM_022353
|
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1
|
|
chr11_-_117628211
|
3.458
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr3_-_172660545
|
3.399
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr2_-_45691887
|
3.390
|
NM_018079
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr16_+_23560187
|
3.390
|
NM_001199011
NM_032486
|
DCTN5
|
dynactin 5 (p25)
|
|
chr6_+_139136349
|
3.370
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr6_+_33530297
|
3.370
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr8_-_22582526
|
3.362
|
NM_018688
|
BIN3
|
bridging integrator 3
|
|
chr14_-_22127902
|
3.314
|
|
DAD1
|
defender against cell death 1
|
|
chr13_-_36531568
|
3.310
|
NM_001014286
NM_017569
|
FAM48A
|
family with sequence similarity 48, member A
|
|
chr16_-_23559978
|
3.283
|
NM_024675
|
PALB2
|
partner and localizer of BRCA2
|
|
chr2_-_68332985
|
3.281
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr4_+_109761170
|
3.280
|
NM_000995
NM_033625
|
RPL34
|
ribosomal protein L34
|
|
chr2_-_175741096
|
3.273
|
NM_001880
|
ATF2
|
activating transcription factor 2
|
|
chr11_-_57054756
|
3.267
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr2_-_241690365
|
3.250
|
NM_182501
|
MTERFD2
|
MTERF domain containing 2
|
|
chr17_+_70520339
|
3.217
|
NM_001545
|
ICT1
|
immature colon carcinoma transcript 1
|
|
chr13_+_52124831
|
3.197
|
NM_001130912
NM_006704
|
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae)
|
|
chr2_-_65308220
|
3.195
|
|
LOC729324
|
hCG1986447
|
|
chr20_+_16658608
|
3.195
|
NM_003092
NM_198220
|
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B
|
|
chr16_-_8798973
|
3.193
|
NM_015421
|
TMEM186
|
transmembrane protein 186
|
|
chr7_+_104959767
|
3.186
|
NM_021930
|
RINT1
|
RAD50 interactor 1
|
|
chr1_+_109036431
|
3.172
|
NM_018061
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr17_-_39447817
|
3.168
|
NM_032376
|
TMEM101
|
transmembrane protein 101
|
|
chr3_-_130362563
|
3.165
|
NM_020701
|
ISY1
|
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr7_+_50318801
|
3.165
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr16_+_24648446
|
3.151
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr2_-_20053256
|
3.135
|
NM_001006657
NM_020779
|
WDR35
|
WD repeat domain 35
|
|
chr1_+_220884257
|
3.133
|
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr7_-_91713044
|
3.133
|
NM_004912
NM_001013406
NM_194454
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr3_+_180548173
|
3.022
|
NM_033540
|
MFN1
|
mitofusin 1
|
|
chr12_+_122652572
|
3.000
|
NM_020936
|
DDX55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55
|
|
chr1_-_3702908
|
2.990
|
NM_020710
|
LRRC47
|
leucine rich repeat containing 47
|
|
chr21_+_36614347
|
2.984
|
NM_015358
|
MORC3
|
MORC family CW-type zinc finger 3
|
|
chr20_-_472335
|
2.982
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr1_+_23990228
|
2.981
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr20_+_61760084
|
2.980
|
NM_016434
NM_032957
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr14_-_21049204
|
2.975
|
NM_019852
|
METTL3
|
methyltransferase like 3
|
|
chr3_+_44571600
|
2.972
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr9_-_124733599
|
2.965
|
NM_020924
|
ZBTB26
|
zinc finger and BTB domain containing 26
|
|
chr14_+_23771942
|
2.962
|
NM_001002002
NM_016576
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr11_-_71501385
|
2.953
|
NM_014042
|
C11orf51
|
chromosome 11 open reading frame 51
|
|
chr4_+_110956360
|
2.948
|
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr1_-_53458822
|
2.935
|
NM_017887
|
C1orf123
|
chromosome 1 open reading frame 123
|
|
chr16_-_79668304
|
2.926
|
NM_001100873
NM_152337
|
C16orf46
|
chromosome 16 open reading frame 46
|
|
chr12_-_47698781
|
2.923
|
NM_002733
NM_212461
|
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit
|
|
chr2_+_74535700
|
2.922
|
NM_031288
|
INO80B
|
INO80 complex subunit B
|
|
chr7_+_6583560
|
2.922
|
NM_001134387
NM_001134388
NM_001134389
NM_018106
|
ZDHHC4
|
zinc finger, DHHC-type containing 4
|
|
chr10_-_51293220
|
2.889
|
NM_006327
|
TIMM23
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog (yeast)
translocase of inner mitochondrial membrane 23 homolog B (yeast)
|
|
chr10_-_51041156
|
2.861
|
NM_003631
|
AGAP8
LOC728407
PARG
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8
poly(ADP-ribose) glycohydrolase pseudogene
poly (ADP-ribose) glycohydrolase
|
|
chr5_+_69381105
|
2.860
|
NM_000344
NM_022874
NM_017411
NM_022875
NM_022876
NM_022877
|
SMN1
SMN2
|
survival of motor neuron 1, telomeric
survival of motor neuron 2, centromeric
|
|
chr4_+_56509793
|
2.837
|
NM_025009
|
CEP135
|
centrosomal protein 135kDa
|
|
chr2_+_239000395
|
2.833
|
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chr2_-_131567458
|
2.825
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr1_+_39229474
|
2.823
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr3_+_32701663
|
2.798
|
NM_015442
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr6_-_10802823
|
2.797
|
NM_001145020
|
C6orf52
|
chromosome 6 open reading frame 52
|
|
chr16_-_70400232
|
2.792
|
|
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit
|
|
chr2_+_48395271
|
2.789
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr12_+_56424930
|
2.770
|
NM_005981
|
TSPAN31
|
tetraspanin 31
|
|
chr10_-_18980512
|
2.759
|
NM_182543
|
NSUN6
|
NOP2/Sun domain family, member 6
|
|
chr20_-_57040700
|
2.757
|
NM_006886
NM_001001977
|
ATP5E
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit
|
|
chr4_-_13238368
|
2.742
|
NM_148894
|
BOD1L
|
biorientation of chromosomes in cell division 1-like
|
|
chr5_-_159778621
|
2.734
|
NM_006425
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae)
|
|
chr7_+_39572396
|
2.730
|
NM_020192
|
C7orf36
|
chromosome 7 open reading frame 36
|
|
chr9_-_133141726
|
2.729
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr9_+_36562844
|
2.726
|
NM_014791
|
MELK
|
maternal embryonic leucine zipper kinase
|
|
chr3_+_144203061
|
2.716
|
NM_001080415
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr5_+_74098828
|
2.715
|
NM_014886
|
NSA2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae)
|
|
chr2_+_118562499
|
2.714
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chr6_+_41714171
|
2.711
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr7_-_32496497
|
2.706
|
NM_012322
|
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr1_+_229731021
|
2.682
|
NM_005999
|
TSNAX-DISC1
TSNAX
|
TSNAX-DISC1 read-through gene
translin-associated factor X
|
|
chr1_+_45926433
|
2.677
|
NM_016486
|
TMEM69
|
transmembrane protein 69
|
|
chr2_+_113058642
|
2.670
|
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr8_-_120914233
|
2.663
|
NM_003184
|
TAF2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa
|
|
chr11_-_13441294
|
2.653
|
NM_032320
|
BTBD10
|
BTB (POZ) domain containing 10
|
|
chr2_+_27659396
|
2.653
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr2_+_3362449
|
2.644
|
NM_016030
|
TTC15
|
tetratricopeptide repeat domain 15
|
|
chr1_+_172104115
|
2.637
|
NM_001122770
NM_032522
|
ZBTB37
|
zinc finger and BTB domain containing 37
|
|
chr17_-_74990157
|
2.635
|
NM_001082575
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr14_-_38971370
|
2.634
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr1_-_32888677
|
2.631
|
NM_178547
|
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand
|
|
chr2_+_100985122
|
2.627
|
NM_000993
NM_001098577
NM_001099693
|
RPL31
|
ribosomal protein L31
|
|
chr2_+_27740254
|
2.601
|
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein
|
|
chr11_+_8660913
|
2.598
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr8_+_11179724
|
2.593
|
|
MTMR9
|
myotubularin related protein 9
|
|
chr12_+_49080858
|
2.562
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr2_-_240724373
|
2.557
|
NM_001163424
NM_138336
|
MYEOV2
|
myeloma overexpressed 2
|
|
chr16_+_2672527
|
2.548
|
|
KCTD5
|
potassium channel tetramerisation domain containing 5
|
|
chr2_+_122124034
|
2.545
|
|
LOC254128
|
hypothetical LOC254128
|
|
chr2_-_197883569
|
2.544
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr1_+_212520899
|
2.519
|
NM_020197
|
SMYD2
|
SET and MYND domain containing 2
|
|
chr4_-_84595967
|
2.517
|
NM_133636
|
HELQ
|
helicase, POLQ-like
|
|
chr14_-_22127954
|
2.513
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chr14_+_22368942
|
2.509
|
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr20_-_5879036
|
2.507
|
NM_015939
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae)
|
|
chr16_-_82778071
|
2.506
|
NM_005679
NM_139353
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa
|
|
chr14_+_20527971
|
2.506
|
|
METTL17
|
methyltransferase like 17
|
|
chr6_+_32034559
|
2.502
|
NM_006929
|
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae)
|
|
chr17_-_975809
|
2.499
|
|
ABR
|
active BCR-related gene
|
|
chr4_+_84596141
|
2.498
|
NM_016067
|
MRPS18C
|
mitochondrial ribosomal protein S18C
|
|
chr15_+_48503860
|
2.492
|
NM_001128610
NM_001128611
NM_005154
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr20_-_31452991
|
2.474
|
NM_016082
NM_016408
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr11_+_61647990
|
2.474
|
NM_001040694
NM_020238
|
INCENP
|
inner centromere protein antigens 135/155kDa
|
|
chr1_-_38228179
|
2.468
|
NM_006802
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa
|
|
chr17_+_42355492
|
2.454
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr17_+_35629080
|
2.453
|
NM_133264
|
WIPF2
|
WAS/WASL interacting protein family, member 2
|
|
chr14_-_23981493
|
2.452
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr20_+_47862782
|
2.439
|
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr2_+_98591516
|
2.437
|
|
UNC50
|
unc-50 homolog (C. elegans)
|